
Cicero predicts cis-regulatory dna interactions from single-cell chromatin accessibility data. Dimensionality reduction for visualizing single-cell data using UMAP. Spatial reconstruction of single-cell gene expression data. Coupled single-cell CRISPR screening and epigenomic profiling reveals causal gene regulatory networks. Organoid modeling of the tumor immune microenvironment. Massively parallel digital transcriptional profiling of single cells. A single-cell atlas of in vivo mammalian chromatin accessibility. Integrated single-cell analysis maps the continuous regulatory landscape of human hematopoietic differentiation. Transcript-indexed ATAC-seq for precision immune profiling. The cis-regulatory dynamics of embryonic development at single-cell resolution. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution.
Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Single-cell chromatin accessibility reveals principles of regulatory variation. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data.
Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Integrative analysis of 111 reference human epigenomes. We anticipate that scATAC-seq will enable the unbiased discovery of gene regulatory factors across diverse biological systems.
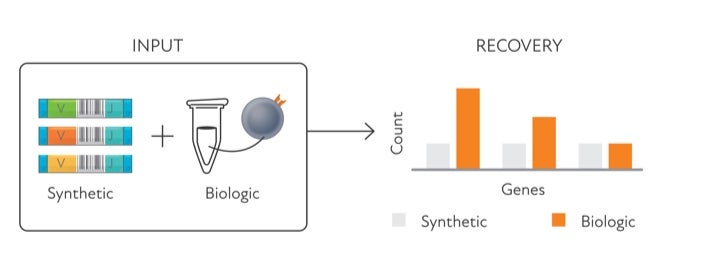
#IMMUNE REPERTOIRE CAPTURE STANFORD NATURE BIOTECH SERIAL#
Analysis of scATAC-seq profiles from serial tumor biopsies before and after programmed cell death protein 1 blockade identifies chromatin regulators of therapy-responsive T cell subsets and reveals a shared regulatory program that governs intratumoral CD8 + T cell exhaustion and CD4 + T follicular helper cell development. In basal cell carcinoma, application of scATAC-seq reveals regulatory networks in malignant, stromal and immune cells in the tumor microenvironment. In blood, application of scATAC-seq enables marker-free identification of cell type-specific cis- and trans-regulatory elements, mapping of disease-associated enhancer activity and reconstruction of trajectories of cellular differentiation. We apply scATAC-seq to obtain chromatin profiles of more than 200,000 single cells in human blood and basal cell carcinoma. Here, we assess the performance of a massively parallel droplet-based method for mapping transposase-accessible chromatin in single cells using sequencing (scATAC-seq).
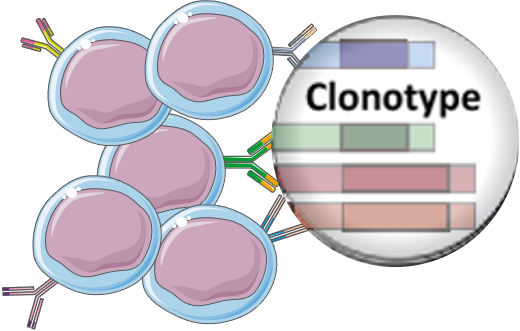
This unique tool is based on T-cell receptor sequencing data generated from Adaptive’s partnership with Microsoft and our Antigen Map project from ~4,000 HLA typed samples.Understanding complex tissues requires single-cell deconstruction of gene regulation with precision and scale. The HLA classifier uses machine learning to provide a positive or negative call for 145 HLA genes and alleles. The Immunosequencing assay can infer a sample’s HLA type based on the T-cell receptor (TCR) profile. By adding the HLA classifier to samples processed using Immunosequencing, researchers can unlock new understanding of immune repertoire dynamics in the context of HLA type. New research is emerging that explores the link between HLA and disease susceptibility, response to immunotherapy, and more. The importance of a person’s HLA type in transplant research is well understood, as is the association of specific HLA alleles to common diseases. HLA type plays an important role in driving T-cell selection and in shaping T-cell repertoires. Foreign antigens are presented to T cells by protein complexes called “major histocompatibility complexes” (MHC) that are coded by human leukocyte antigen (HLA) genes.


 0 kommentar(er)
0 kommentar(er)
